Human LIMPII/SR-B2 Antibody Summary
Arg27-Thr432
Accession # Q14108
Applications
Human LIMPII/SR-B2 Sandwich Immunoassay
Please Note: Optimal dilutions should be determined by each laboratory for each application. General Protocols are available in the Technical Information section on our website.
Scientific Data
 View Larger
View Larger
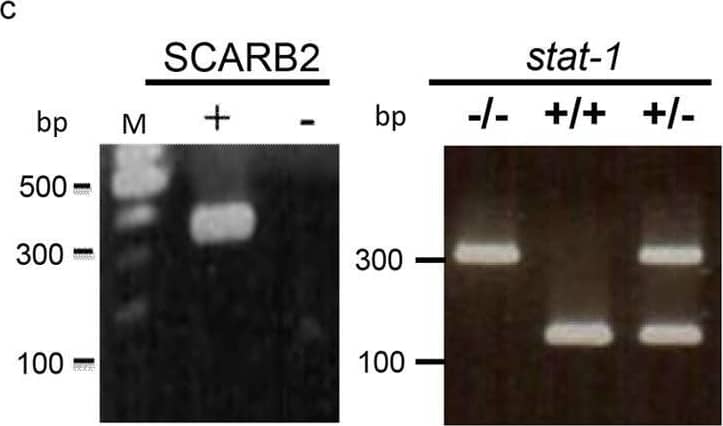
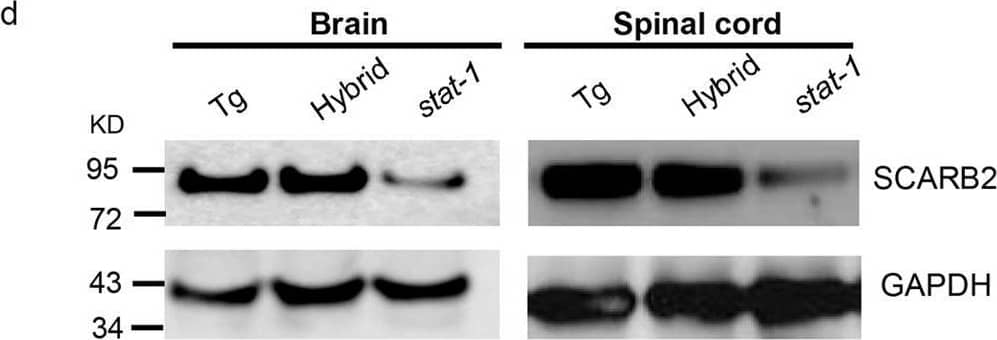
Detection of LIMPII/SR-B2 by Western Blot Generation of a hybrid mouse model SCARB2/stat-1 KO by crossbreeding hSCARB2 transgenic mice and stat-1 KO mice.(a) Human SCARB2 (hSCARB2) cDNA was cloned under its own native promoter in an SV40 expression vector. (b) The generation of homozygous hSCARB2+/+ transgenic mice was as detailed in M&M. The heterozygote strain of hSCARB2+/−/stat-1+/− mice were generated by crossing the stat-1−/− and the hSCARB2+/+ parental mice. The hybrid strain hSCARB2+/+/stat-1−/− was generated by crossing the heterozygote mice hSCARB2+/−/stat-1+/− to each other. (c) Genotyping of parental and hybrid mouse strains was performed by PCR assay using genomic DNAs extracted from mouse tail. The transgene of hSCARB2 was screened by detection of a 369 bp PCR product using primers specific for hSCARB2. For the stat-1+/− heterozygote mice, 320 bp and 150 bp PCR products were used as markers for screening. The former indicates a mutant stat-1 allele, while the latter indicates a wild type stat-1 allele. (d) The expressions of hSCARB2 protein were compared among parental and hybrid mouse strains in brain, spinal cord, spleen and muscle by immunoblot via an anti-SCARB2 antibody. The weaker signals of the SCARB2 protein detected in stat-1 KO mice reflect cross reactivity between human and mouse SCARB2 proteins to the anti-SCARB2 antibody. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/27499235), licensed under a CC-BY license. Not internally tested by R&D Systems.
 View Larger
View Larger
Detection of LIMPII/SR-B2 by Western Blot Generation of a hybrid mouse model SCARB2/stat-1 KO by crossbreeding hSCARB2 transgenic mice and stat-1 KO mice.(a) Human SCARB2 (hSCARB2) cDNA was cloned under its own native promoter in an SV40 expression vector. (b) The generation of homozygous hSCARB2+/+ transgenic mice was as detailed in M&M. The heterozygote strain of hSCARB2+/−/stat-1+/− mice were generated by crossing the stat-1−/− and the hSCARB2+/+ parental mice. The hybrid strain hSCARB2+/+/stat-1−/− was generated by crossing the heterozygote mice hSCARB2+/−/stat-1+/− to each other. (c) Genotyping of parental and hybrid mouse strains was performed by PCR assay using genomic DNAs extracted from mouse tail. The transgene of hSCARB2 was screened by detection of a 369 bp PCR product using primers specific for hSCARB2. For the stat-1+/− heterozygote mice, 320 bp and 150 bp PCR products were used as markers for screening. The former indicates a mutant stat-1 allele, while the latter indicates a wild type stat-1 allele. (d) The expressions of hSCARB2 protein were compared among parental and hybrid mouse strains in brain, spinal cord, spleen and muscle by immunoblot via an anti-SCARB2 antibody. The weaker signals of the SCARB2 protein detected in stat-1 KO mice reflect cross reactivity between human and mouse SCARB2 proteins to the anti-SCARB2 antibody. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/27499235), licensed under a CC-BY license. Not internally tested by R&D Systems.
 View Larger
View Larger
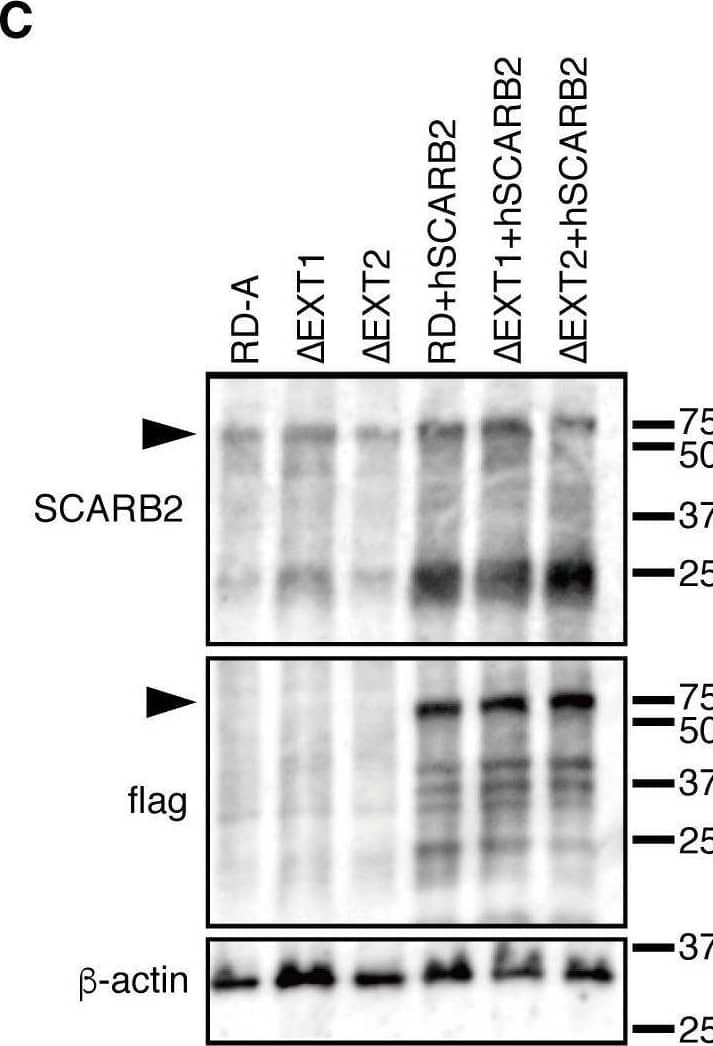
Detection of LIMPII/SR-B2 by Western Blot HS and SCARB2 expression in genetically modified RD-A cells.(A–B) FACS analysis of HS expression at the cell surface. (C) Western blotting analysis using anti-SCARB2 (top), anti-flag (middle), and anti-beta -actin (bottom) antibodies. The arrowheads indicate hSCARB2. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/32187235), licensed under a CC-BY license. Not internally tested by R&D Systems.
 View Larger
View Larger
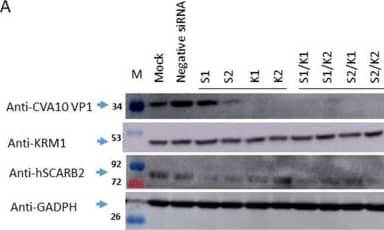
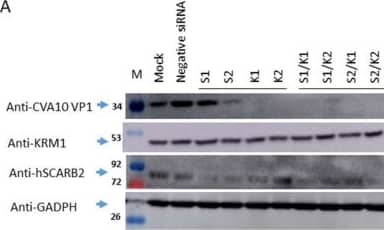
Detection of LIMPII/SR-B2 by Western Blot The reduction of endogenous hSCARB2 and KRM1 expression impacted CVA10 infection. RD Cells were transfected individually with 50 pmoles of siRNA specific to hSCARB2 (s2651(S1) and s2652(S2)), KRM1 (s38393(K1) and s38394(K2)), or mixed specific siRNA with equal amounts (50 + 50 pmoles) of S1 + K1, S1 + K2, S2 + K1, S2 + K2, or negative siRNA, followed by 48 h of incubation. Infection of siRNA-treated cells with CVA10 (MOI = 0.05) and then cultured for another 24 h then proceeded. (a) Western blotting with the respective antibody examined the expression level of hSCARB2 or KRN1 in the cells. (b) The supernatant and (c) lysate were collected and subjected to a plaque-forming assay to detect the amounts of produced CVA10, and the results were shown. (d) Total amounts of CVA10 production using the sum of the viral amounts from (b,c) are shown. The symbols **, *** and **** were used to indicate p < 0.01, p < 0.001, and p < 0.0001, respectively. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/37112912), licensed under a CC-BY license. Not internally tested by R&D Systems.
 View Larger
View Larger
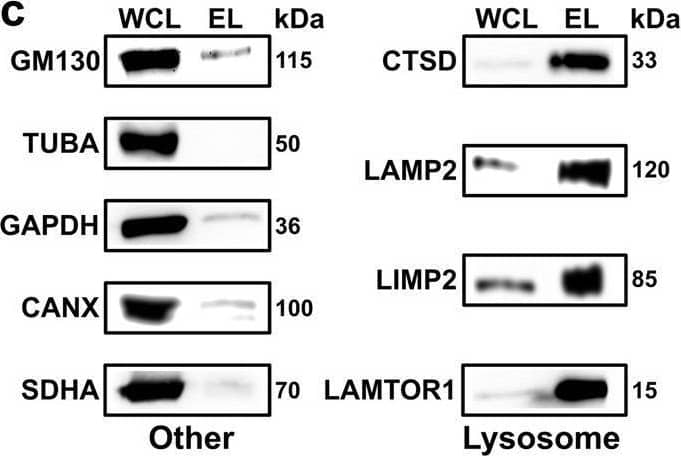
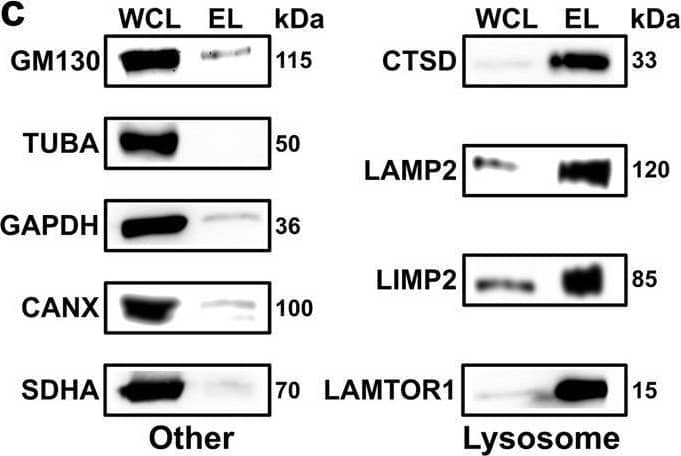
Detection of LIMPII/SR-B2 by Western Blot Cross-linking mass spectrometry analysis of lysosome-enriched fractions. A Experimental workflow for the XL-LC-MS/MS analysis of lysosome-enriched fractions. Created with BioRender.com. b Normalized beta -hexosaminidase activities for individual fractions from lysosome enrichment by SPIONs. Data are presented as mean values + SD (n = 3, biologically independent samples over three independent experiments). c Western blot analysis of lysosome-enriched fractions for contamination by other organelles (n = 2). Lysosome: lysosomal proteins (CTSD, LAMP2, LIMP2, and LAMTOR1). Other: Golgi apparatus (GM130), cytoskeleton (TUBA), cytosol (GAPDH), endoplasmic reticulum (CANX), and mitochondria (SDHA). d Summed iBAQ abundances for proteins identified in lysosome-enriched fractions in ≥3 replicates. e Classification of unique cross-linked residue pairs. f Proteins detected in non-cross-linked lysosome-enriched fractions (proteome), and unique lysosomal cross-linked residue pairs (interactome) for DR and IT samples. g Localization of CSMs for 68 lysosomal proteins cross-linked in the DR and IT state. Cytosolic: proteins located at the cytosolic face of the lysosomal membrane; Lumen: lysosomal luminal proteins. h Correlation of cross-link identification and protein abundance for lysosomal proteins. CSMs and PSMs represent summed values of the analysis (n = 6, biologically independent samples over six independent experiments (3× DR and 3× IT)). SPIONs superparamagnetic iron oxide nanoparticles, DR disrupted, IT intact, SCX strong cation-exchange, IN input, FT flow through, W wash, EL eluate, WCL whole-cell lysate, iBAQ intensity-based absolute quantification, XLs cross-links, CSMs cross-link spectral matches, PSMs peptide spectral matches. Source data are provided as a Source Data file. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/36266287), licensed under a CC-BY license. Not internally tested by R&D Systems.
 View Larger
View Larger
Detection of LIMPII/SR-B2 by Western Blot The reduction of endogenous hSCARB2 and KRM1 expression impacted CVA10 infection. RD Cells were transfected individually with 50 pmoles of siRNA specific to hSCARB2 (s2651(S1) and s2652(S2)), KRM1 (s38393(K1) and s38394(K2)), or mixed specific siRNA with equal amounts (50 + 50 pmoles) of S1 + K1, S1 + K2, S2 + K1, S2 + K2, or negative siRNA, followed by 48 h of incubation. Infection of siRNA-treated cells with CVA10 (MOI = 0.05) and then cultured for another 24 h then proceeded. (a) Western blotting with the respective antibody examined the expression level of hSCARB2 or KRN1 in the cells. (b) The supernatant and (c) lysate were collected and subjected to a plaque-forming assay to detect the amounts of produced CVA10, and the results were shown. (d) Total amounts of CVA10 production using the sum of the viral amounts from (b,c) are shown. The symbols **, *** and **** were used to indicate p < 0.01, p < 0.001, and p < 0.0001, respectively. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/37112912), licensed under a CC-BY license. Not internally tested by R&D Systems.
 View Larger
View Larger
Detection of LIMPII/SR-B2 by Western Blot Cross-linking mass spectrometry analysis of lysosome-enriched fractions. A Experimental workflow for the XL-LC-MS/MS analysis of lysosome-enriched fractions. Created with BioRender.com. b Normalized beta -hexosaminidase activities for individual fractions from lysosome enrichment by SPIONs. Data are presented as mean values + SD (n = 3, biologically independent samples over three independent experiments). c Western blot analysis of lysosome-enriched fractions for contamination by other organelles (n = 2). Lysosome: lysosomal proteins (CTSD, LAMP2, LIMP2, and LAMTOR1). Other: Golgi apparatus (GM130), cytoskeleton (TUBA), cytosol (GAPDH), endoplasmic reticulum (CANX), and mitochondria (SDHA). d Summed iBAQ abundances for proteins identified in lysosome-enriched fractions in ≥3 replicates. e Classification of unique cross-linked residue pairs. f Proteins detected in non-cross-linked lysosome-enriched fractions (proteome), and unique lysosomal cross-linked residue pairs (interactome) for DR and IT samples. g Localization of CSMs for 68 lysosomal proteins cross-linked in the DR and IT state. Cytosolic: proteins located at the cytosolic face of the lysosomal membrane; Lumen: lysosomal luminal proteins. h Correlation of cross-link identification and protein abundance for lysosomal proteins. CSMs and PSMs represent summed values of the analysis (n = 6, biologically independent samples over six independent experiments (3× DR and 3× IT)). SPIONs superparamagnetic iron oxide nanoparticles, DR disrupted, IT intact, SCX strong cation-exchange, IN input, FT flow through, W wash, EL eluate, WCL whole-cell lysate, iBAQ intensity-based absolute quantification, XLs cross-links, CSMs cross-link spectral matches, PSMs peptide spectral matches. Source data are provided as a Source Data file. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/36266287), licensed under a CC-BY license. Not internally tested by R&D Systems.
Reconstitution Calculator
Preparation and Storage
- 12 months from date of receipt, -20 to -70 °C as supplied.
- 1 month, 2 to 8 °C under sterile conditions after reconstitution.
- 6 months, -20 to -70 °C under sterile conditions after reconstitution.
Background: LIMPII/SR-B2
LIMPII (Lysosomal Integral Membrane Protein II), also known as LPG85 (85 kDa lysosomal membrane sialoglycoprotein) and as CD36 antigen-like 2 (CD36L2), is a major lysosomal membrane protein. It belongs to the scavenger receptor class B subfamily and is designated member 2 (SR-B2). Other mammalian members of this family include SR-B1 (alternatively known as Cla-1 and CD36L1), and SR-B3 (CD36) (1 - 3). SR-B/CD36 family members are type III integral membrane proteins with an N- as well as a C-terminal cytoplasmic tail, and a large extracellular (or lumenal in the case of LIMPII) loop containing similarly spaced cysteine residues and multiple glycosylation sites. The C-terminal cytoplasmic tail has a di-leucine-based motif that mediates effective lysosomal targeting. LIMPII is widely expressed on all tissues and cell types so far examined. It is also expressed on the surface of activated platelets. LIMPII binds thrombospondin-1, but the biological significance of this interaction is not known. LIMPII-thrombospondin interaction may contribute to the pro-adhesive changes of activated platelets during coagulation, and inflammation (1). Overexpression of LIMPII causes an enlargement of early endosomes and late endosomes, suggesting that LIMPII may play a role in lysosome/endosome biogenesis (4). Mice deficient in LIMPII are impaired in membrane transport processes, resulting in ureteric pelvic junction obstruction, deafness and peripheral neuropathy (5).
- Crombie, R. and R. Silverstein (1998) J. Biol. Chem. 273:4855.
- Febbraio, M. et al. (2001) J. Clin. Invest. 108:785.
- Eskelinen, E-L. et al. (2003) Trends in Cell Biology 13:137.
- Kuronita, T. et al. (2002) J. Cell Sci. 115:4117.
- Gamp, A-C. et al. (2003) Human Molecular Genetics 12:631.
Product Datasheets
Citations for Human LIMPII/SR-B2 Antibody
R&D Systems personnel manually curate a database that contains references using R&D Systems products. The data collected includes not only links to publications in PubMed, but also provides information about sample types, species, and experimental conditions.
17
Citations: Showing 1 - 10
Filter your results:
Filter by:
-
The uncoating of EV71 in mature late endosomes requires CD-M6PR
Authors: Seii Ohka, Soon Hao Tan, Eri Ishiyama, Katsutoshi Ogasawara, Tomohito Hanasaka, Kinji Ishida et al.
Biology Open
-
A Novel Murine Model Expressing a Chimeric mSCARB2/hSCARB2 Receptor Is Highly Susceptible to Oral Infection with Clinical Isolates of Enterovirus 71
Authors: Cheng-Hung Yang, Chung-Tiang Liang, Si-Tse Jiang, Kuan-Hsing Chen, Chun-Chiao Yang, Mei-Ling Cheng et al.
Journal of Virology
-
Pathogenesis Study of Enterovirus 71 Using a Novel Human SCARB2 Knock-In Mouse Model
Authors: Yuefei Jin, Tiantian Sun, Guangyuan Zhou, Dong Li, Shuaiyin Chen, Weiguo Zhang et al.
mSphere
-
Human SCARB2 Acts as a Cellular Associator for Helping Coxsackieviruses A10 Infection
Authors: SL Yu, NH Chung, YC Lin, YA Liao, YC Chen, YH Chow
Viruses, 2023-04-08;15(4):.
Species: Human
Sample Types: Cell Lysates
Applications: Immunoprecipitation -
Human SCARB2 Acts as a Cellular Associator for Helping Coxsackieviruses A10 Infection
Authors: SL Yu, NH Chung, YC Lin, YA Liao, YC Chen, YH Chow
Viruses, 2023;15(4):.
Species: Human
Sample Types: Cell Lysates
Applications: Immunoprecipitation -
Cross-linking of the endolysosomal system reveals potential flotillin structures and cargo
Authors: J Singh, H Elhabashy, P Muthukotti, M Stepath, M Eisenacher, O Kohlbacher, V Gieselmann, D Winter
Nature Communications, 2022-10-20;13(1):6212.
Species: Human
Sample Types: Cell Lysates
Applications: Western Blot -
Proteomics of Primary Uveal Melanoma: Insights into Metastasis and Protein Biomarkers
Authors: GF Jang, JS Crabb, B Hu, B Willard, H Kalirai, AD Singh, SE Coupland, JW Crabb
Cancers, 2021-07-14;13(14):.
Species: Human
Sample Types: Tissue Homogenates
-
Pathogenesis Study of Enterovirus 71 Using a Novel Human SCARB2 Knock-In Mouse Model
Authors: Yuefei Jin, Tiantian Sun, Guangyuan Zhou, Dong Li, Shuaiyin Chen, Weiguo Zhang et al.
mSphere
Species: Mouse
Sample Types: Whole Tissue
Applications: Immunohistochemistry -
Involvement of VCP/UFD1/Nucleolin in the viral entry of Enterovirus A species
Authors: J Yan, M Wang, M Wang, Y Dun, L Zhu, Z Yi, S Zhang
Virus Res., 2020-04-11;283(0):197974.
Species: Human
Sample Types: Cell Lysates
Applications: Western Blot -
Heparan sulfate attachment receptor is a major selection factor for attenuated enterovirus 71 mutants during cell culture adaptation
Authors: K Kobayashi, K Mizuta, S Koike
PLoS Pathog., 2020-03-18;16(3):e1008428.
Species: Human
Sample Types: Cell Lysates
Applications: Western Blot -
Fibronectin facilitates EV71 infection by mediating its entry
Authors: QQ He, S Ren, ZC Xia, ZK Cheng, NF Peng, Y Zhu
J. Virol., 2018-04-13;0(0):.
Species: Human
Sample Types: Whole Cells
Applications: Neutralization -
The binding of a monoclonal antibody to the apical region of SCARB2 blocks EV71 infection
Authors: X Zhang, P Yang, N Wang, J Zhang, J Li, H Guo, X Yin, Z Rao, X Wang, L Zhang
Protein Cell, 2017-04-26;0(0):.
Species: Human, Mouse
Sample Types: Cell Lysates
Applications: Western Blot -
Transcriptome analysis reveals dynamic changes in coxsackievirus A16 infected HEK 293T cells
Authors: J Jin, R Li, C Jiang, R Zhang, X Ge, F Liang, X Sheng, W Dai, M Chen, J Wu, J Xiao, W Su
BMC Genomics, 2017-01-25;18(0):933.
Species: Human
Sample Types: Cell Lysates
Applications: Western Blot -
Amphotericin B Inhibits Enterovirus 71 Replication by Impeding Viral Entry
Sci Rep, 2016-09-09;6(0):33150.
Species: Human
Sample Types: Cell Lysates
Applications: Western Blot -
The role of Misshapen NCK-related kinase (MINK), a novel Ste20 family kinase, in the IRES-mediated protein translation of human enterovirus 71.
Authors: Leong, Shi Yun, Ong, Bryan Ki, Chu, Justin J
PLoS Pathog, 2015-03-06;11(3):e1004686.
Species: Human
Sample Types: Whole Cells
Applications: Functional Assay -
Scavenger receptor B2 is a cellular receptor for enterovirus 71.
Authors: Yamayoshi S, Yamashita Y, Li J, Hanagata N, Minowa T, Takemura T, Koike S
Nat. Med., 2009-06-21;15(7):798-801.
Species: Human
Sample Types: Whole Cells
Applications: Flow Cytometry, Neutralization -
All three human scavenger receptor class B proteins can bind and transport all three macular xanthophyll carotenoids
Authors: Rajalekshmy Shyam, Preejith Vachali, Aruna Gorusupudi, Kelly Nelson, Paul S. Bernstein
Archives of Biochemistry and Biophysics
FAQs
No product specific FAQs exist for this product, however you may
View all Antibody FAQsReviews for Human LIMPII/SR-B2 Antibody
There are currently no reviews for this product. Be the first to review Human LIMPII/SR-B2 Antibody and earn rewards!
Have you used Human LIMPII/SR-B2 Antibody?
Submit a review and receive an Amazon gift card.
$25/€18/£15/$25CAN/¥75 Yuan/¥2500 Yen for a review with an image
$10/€7/£6/$10 CAD/¥70 Yuan/¥1110 Yen for a review without an image
