Human XPE/DDB2 Antibody Summary
Met1-Lys427
Accession # Q92466
Applications
Please Note: Optimal dilutions should be determined by each laboratory for each application. General Protocols are available in the Technical Information section on our website.
Scientific Data
 View Larger
View Larger
Detection of Human XPE/DDB2 by Western Blot. Western blot shows lysates of HeLa human cervical epithelial carcinoma cell line and U2OS human osteosarcoma cell line. PVDF membrane was probed with 0.5 µg/mL of Human XPE/DDB2 Antigen Affinity-purified Polyclonal Antibody (Catalog # AF3297) followed by HRP-conjugated Anti-Goat IgG Secondary Antibody (Catalog # HAF017). A specific band was detected for XPE/DDB2 at approximately 45 kDa (as indicated). This experiment was conducted using Immunoblot Buffer Group 1.
 View Larger
View Larger
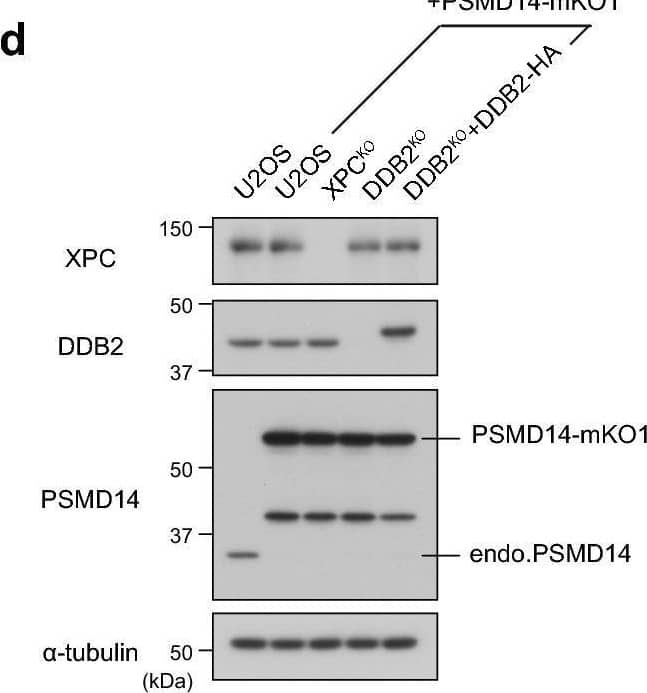
Detection of XPE/DDB2 by Western Blot MG132-induced DDB2 accumulation depends on the proteasome subunit PSMD14. (a) Immunoblot analyses validating depletion of the proteasome subunits. U2OS cells were transfected with negative control siRNA (siNC) or siRNA targeting PSMD14 (siPSMD14), and cell lysates were prepared after incubation for the indicated number of days. All subsequent experiments were performed on the second day of post-transfection. (b,c) Cells expressing DDB2-mKO1 were treated with siNC or siPSMD14, and then incubated for 6 h in the presence or absence of 5 µM MG132. Based on live-cell images acquired with a confocal laser scanning microscope (b), percentages of cells with subnuclear DDB2 accumulation were measured as in Fig. 2d. (d) Immunoblot analyses validating expression of the XPC, DDB2, and PSMD14 proteins in the indicated cell lines. (e) Recruitment of PSMD14-mKO1 to local DNA damage was assessed quantitatively in the indicated cell lines. ** P < 0.01. (f) Representative live-cell images of PSMD14-mKO1 localization in cells that were mock-treated or treated for 6 h in the presence of 5 µM MG132 or 10 µM lactacystin. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/33184426), licensed under a CC-BY license. Not internally tested by R&D Systems.
 View Larger
View Larger
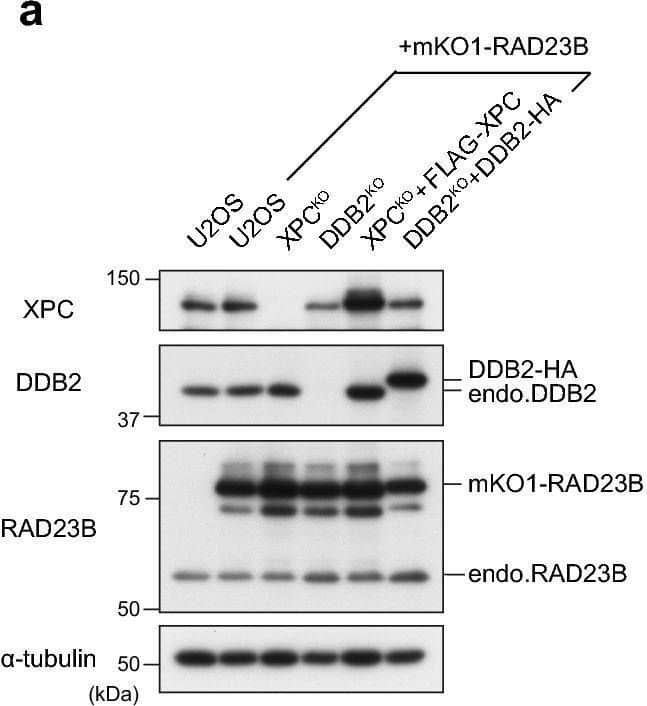
Detection of XPE/DDB2 by Western Blot Real-time monitoring of recruitment of mKO1-RAD23B to local DNA damage. (a) Immunoblot analyses of endogenous and ectopically expressed XPC, DDB2, and RAD23B in the indicated cell lines. (b,c) Local DNA damage was induced in the indicated cell lines by three-photon absorption with an infrared femtosecond laser. Relative mKO1 intensities at damaged sites were quantitatively assessed over time. Inset in (b) shows enlargement of the graph in the early time range. The statistical significance assessed for the last time point is shown. *** P < 0.001. (d) Immunoblot analyses validating the effects of MLN4924. U2OS cells were pre-incubated for 2 h in the presence or absence of 1 µM MLN4924, and then irradiated with UVC at 10 J/m2 (or mock-irradiated) and further incubated for 1 h with or without MLN4924. Asterisk indicates non-specific reaction of the anti-XPC antibody. e The indicated cell lines were pre-treated for 2 h with or without 1 µM MLN4924, and recruitment of mKO1-RAD23B to local DNA damage was measured as in panels b and c. *** P < 0.001. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/33184426), licensed under a CC-BY license. Not internally tested by R&D Systems.
 View Larger
View Larger
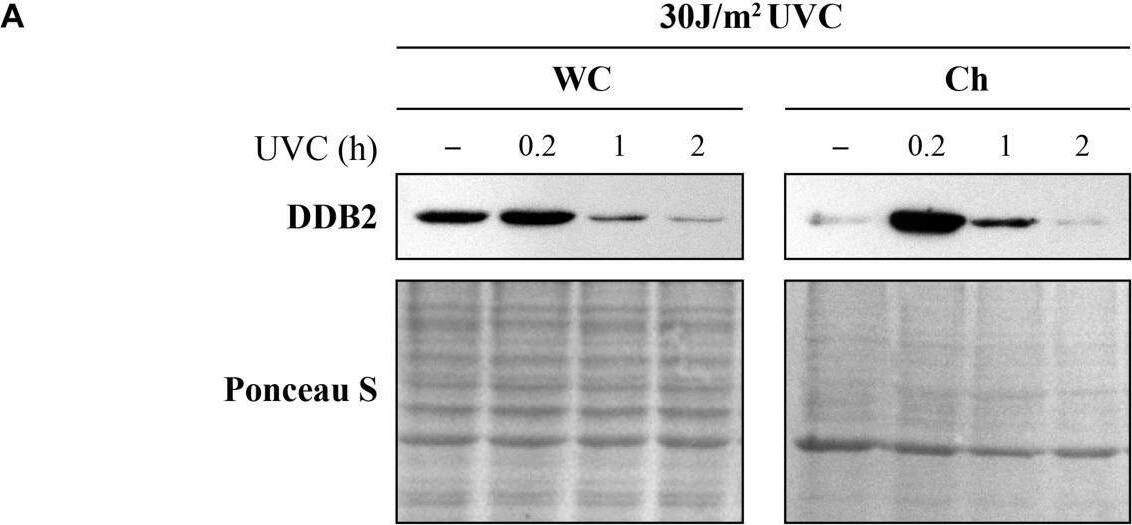
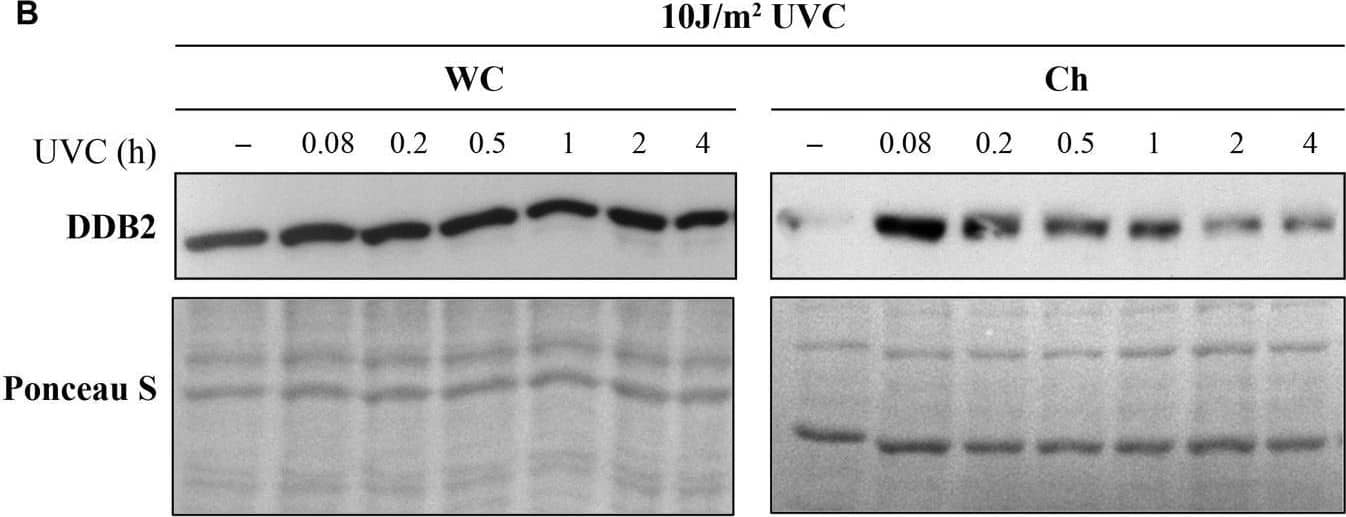
Detection of XPE/DDB2 by Western Blot Ultraviolet radiation irradiation induces the DDB2 ubiquitination and degradation. The GMU6 cells were globally irradiated with UVC at 30 (A) or 10 J/m2(B), and fractionated at the indicated time. The whole cell extracts and chromatin-bound protein fractions were separated on SDS-PAGE and immunoblotted for DDB2. Where specified in panel (B), 10 μM of the proteasome inhibitor MG132 was added 1 h before irradiation with 10 J/m2 UVC to show that the time-dependent depletion of DDB2 in Ch fraction is a result of proteasomal degradation of PTM-modified DDB2. Ponceau S staining was used as loading control. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/33282869), licensed under a CC-BY license. Not internally tested by R&D Systems.
 View Larger
View Larger
Detection of XPE/DDB2 by Western Blot Ultraviolet radiation irradiation induces the DDB2 ubiquitination and degradation. The GMU6 cells were globally irradiated with UVC at 30 (A) or 10 J/m2(B), and fractionated at the indicated time. The whole cell extracts and chromatin-bound protein fractions were separated on SDS-PAGE and immunoblotted for DDB2. Where specified in panel (B), 10 μM of the proteasome inhibitor MG132 was added 1 h before irradiation with 10 J/m2 UVC to show that the time-dependent depletion of DDB2 in Ch fraction is a result of proteasomal degradation of PTM-modified DDB2. Ponceau S staining was used as loading control. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/33282869), licensed under a CC-BY license. Not internally tested by R&D Systems.
 View Larger
View Larger
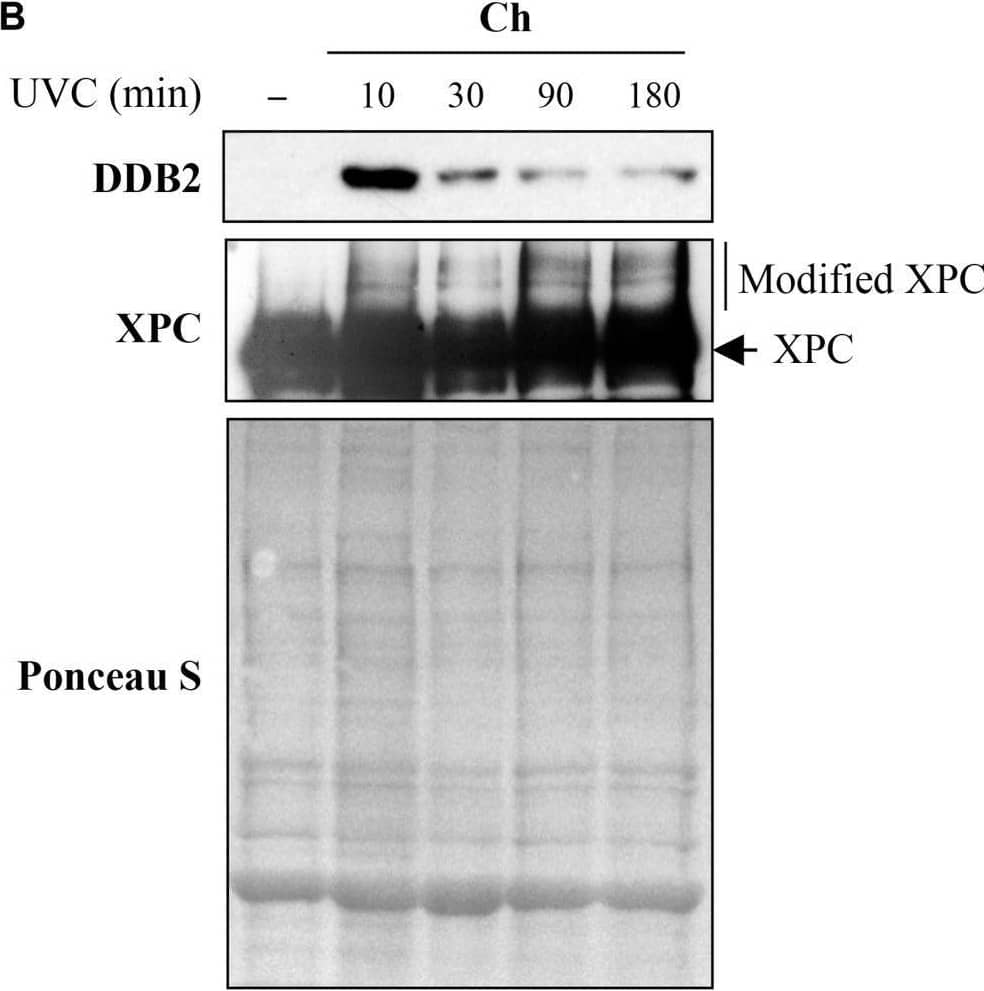
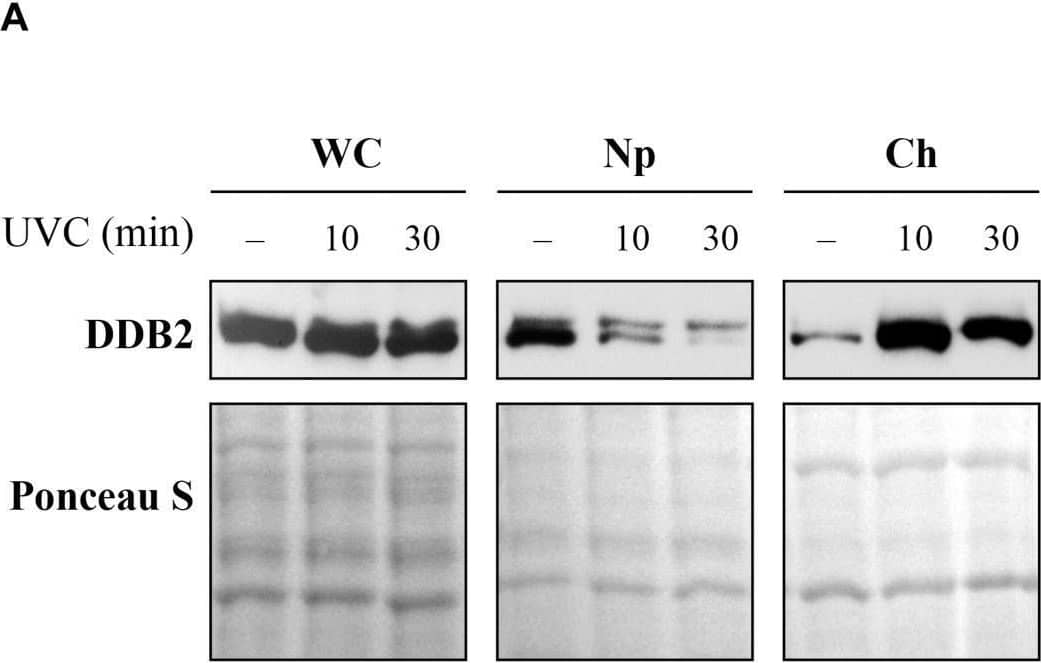
Detection of XPE/DDB2 by Western Blot Sub-cellular and sub-nuclear fractionation of globally UVC-irradiated cells to identify movement and localization of NER proteins to UV-damaged chromatin. The GMU6 cells globally irradiated with 10 J/m2 were fractionated and subjected to Western blot analysis. Ponceau S staining was used as control for loading. (A) The sub-cellular fractions were probed for DDB2 to demonstrate its significant time-dependent depletion in the nucleoplasm and concomitant accumulation in the chromatin-bound protein fraction signifying its movement from inside the nucleus to damaged site on the chromatin in response to global UV-irradiation of the cells. (B) Kinetics of arrival and departure of DDB2 and treatment-dependent post-translational modification of XPC in the chromatin-bound protein fraction extracted at different time points after global UVC irradiation. Ponceau S staining was used as loading control. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/33282869), licensed under a CC-BY license. Not internally tested by R&D Systems.
 View Larger
View Larger
Detection of XPE/DDB2 by Western Blot Sub-cellular and sub-nuclear fractionation of globally UVC-irradiated cells to identify movement and localization of NER proteins to UV-damaged chromatin. The GMU6 cells globally irradiated with 10 J/m2 were fractionated and subjected to Western blot analysis. Ponceau S staining was used as control for loading. (A) The sub-cellular fractions were probed for DDB2 to demonstrate its significant time-dependent depletion in the nucleoplasm and concomitant accumulation in the chromatin-bound protein fraction signifying its movement from inside the nucleus to damaged site on the chromatin in response to global UV-irradiation of the cells. (B) Kinetics of arrival and departure of DDB2 and treatment-dependent post-translational modification of XPC in the chromatin-bound protein fraction extracted at different time points after global UVC irradiation. Ponceau S staining was used as loading control. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/33282869), licensed under a CC-BY license. Not internally tested by R&D Systems.
 View Larger
View Larger
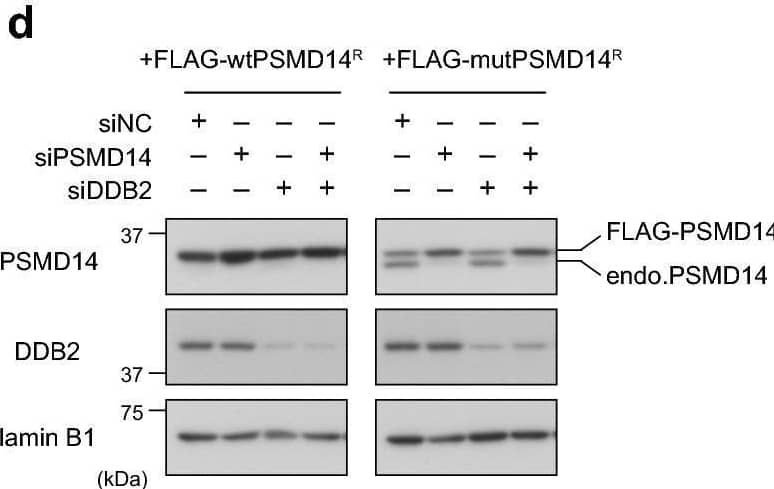
Detection of XPE/DDB2 by Western Blot Presence of a functional proteasome is required for efficient CPD repair. (a,b) U2OS cells treated with siNC or siPSMD14 were used to assess repair kinetics of 6-4PPs (a) and CPDs (b) as in Fig. 4. * P < 0.05. (c) Cells expressing DDB2-mKO1 were treated with siNC or siPSMD14, and recruitment of DDB2-mKO1 to local DNA damage was quantitatively assessed. The statistical significance assessed for the last time point is shown. **** P < 1 × 10–4. (d) Immunoblot analyses validating the effects of siRNAs and the resistance of the ectopically-expressed PSMD14R proteins to the siPSMD14. (e,f) Cells expressing wtPSMD14R or mutPSMD14R were treated with siPSMD14 only (e) or siDDB2 plus siPSMD14 (f). After UVC irradiation at 2 J/m2, repair kinetics of CPDs were assessed. ** P < 0.01. Image collected and cropped by CiteAb from the following open publication (https://pubmed.ncbi.nlm.nih.gov/33184426), licensed under a CC-BY license. Not internally tested by R&D Systems.
Reconstitution Calculator
Preparation and Storage
- 12 months from date of receipt, -20 to -70 °C as supplied.
- 1 month, 2 to 8 °C under sterile conditions after reconstitution.
- 6 months, -20 to -70 °C under sterile conditions after reconstitution.
Background: XPE/DDB2
Xeroderma Pigmentosum complementation group E (XPE) patients are defective in a heterodimer of damage-specific DNA-binding activity (DDB1/DDB2). DDB2 is a 48 kDa protein upregulated in cells exposed to ultraviolet light. XP patients exhibit hypersensitivity to ultraviolet light and have a heightened incidence of skin cancer.
Product Datasheets
Citations for Human XPE/DDB2 Antibody
R&D Systems personnel manually curate a database that contains references using R&D Systems products. The data collected includes not only links to publications in PubMed, but also provides information about sample types, species, and experimental conditions.
7
Citations: Showing 1 - 7
Filter your results:
Filter by:
-
Fluorescence-microscopy-based assay assessing regulatory mechanisms of global genome nucleotide excision repair in cultured cells
Authors: Masayuki Kusakabe, Kaoru Sugasawa
STAR Protocols
-
XPC-PARP complexes engage the chromatin remodeler ALC1 to catalyze global genome DNA damage repair
Authors: C Blessing, K Apelt, D van den He, C Gonzalez-L, MB Rother, M van der Wo, R González-P, A Yifrach, A Parnas, RG Shah, TT Kuo, DEC Boer, J Cai, A Kragten, HS Kim, OD Schärer, ACO Vertegaal, GM Shah, S Adar, H Lans, H van Attiku, AG Ladurner, MS Luijsterbu
Nature Communications, 2022-08-13;13(1):4762.
Species: Human
Sample Types: Cell Lysates
Applications: Western Blot -
Histone deacetylation regulates nucleotide excision repair through an interaction with the XPC protein
Authors: Masayuki Kusakabe, Erina Kakumu, Fumika Kurihara, Kazuki Tsuchida, Takumi Maeda, Haruto Tada et al.
iScience
-
Functional impacts of the ubiquitin-proteasome system on DNA damage recognition in global genome nucleotide excision repair
Authors: W Sakai, M Yuasa-Suna, M Kusakabe, A Kishimoto, T Matsui, Y Kaneko, JI Akagi, N Huyghe, M Ikura, T Ikura, F Hanaoka, M Yokoi, K Sugasawa
Sci Rep, 2020-11-12;10(1):19704.
Species: Human
Sample Types: Whole Cells
Applications: ICC -
Regulation of liver receptor homologue-1 by DDB2 E3 ligase activity is critical for hepatic glucose metabolism
Authors: TC Lai, MC Hu
Sci Rep, 2019-03-28;9(1):5304.
Species: Human
Sample Types: Cell Lysates
Applications: Western Blot -
Human CRL4DDB2 ubiquitin ligase preferentially regulates post-repair chromatin restoration of H3K56Ac through recruitment of histone chaperon CAF-1
Authors: Q Zhu, S Wei, N Sharma, G Wani, J He, AA Wani
Oncotarget, 2017-10-17;8(61):104525-104542.
Species: Human
Sample Types: Cell Lysates
Applications: Western Blot -
DDB2 increases radioresistance of NSCLC cells by enhancing DNA damage responses
Authors: Ning Zou, Guozhen Xie, Tiantian Cui, Amit Kumar Srivastava, Meihua Qu, Linlin Yang et al.
Tumor Biology
FAQs
No product specific FAQs exist for this product, however you may
View all Antibody FAQsReviews for Human XPE/DDB2 Antibody
There are currently no reviews for this product. Be the first to review Human XPE/DDB2 Antibody and earn rewards!
Have you used Human XPE/DDB2 Antibody?
Submit a review and receive an Amazon gift card.
$25/€18/£15/$25CAN/¥75 Yuan/¥2500 Yen for a review with an image
$10/€7/£6/$10 CAD/¥70 Yuan/¥1110 Yen for a review without an image
