Torin 1
Tocris Bioscience | Catalog # 4247

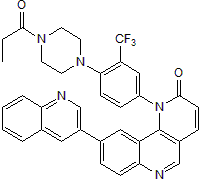
Product Description
Torin 1 is a potent and selective ATP-competitive mTOR inhibitor (IC50 = 2 - 10 nM for mTORC1 and mTORC2). Torin 1 displays 200-fold selectivity for mTOR over DNA-PK, ATM and hVps34. In HeLa cells, Torin 1 induces autophagy. Torin 1 impairs cell growth and proliferation by suppression of the rapamycin-resistant functions of mTORC1. In human monocytes and myeloid dendritic cells, Torin 1 prevents decreases the anti-inflammatory potency of glucocorticoids. In the human endocrine cell line BON, Torin 1 increases neurotensin secretion and gene expression through MEK/ERK/c-Jun pathway activation.Licensing Information
Sold under license from Whitehead Institute for Biomedical Research.
Product Specifications for Torin 1
Molecular Weight
Formula
Storage
Purity
Chemical Name
CAS Number
PubChem ID
InChI Key
SMILES
The technical data provided above is for guidance only. For batch specific data refer to the Certificate of Analysis.
Solubility
| Solvent | Max Conc. mg/mL | Max Conc. mM | |
|---|---|---|---|
| Solubility | |||
| DMSO | 0.61 | 1 with gentle warming |
Preparing Stock Solutions for Torin 1
The following data is based on the product molecular weight 607.62.
Batch specific molecular weights may vary from batch to batch due to the degree of hydration, which all affect the solvent volumes required to prepare stock solutions.
| Concentration / Solvent Volume / Mass | 1 mg | 5 mg | 10 mg |
|---|---|---|---|
| 0.01 mM | 164.58 mL | 822.88 mL | 1,645.77 mL |
| 0.05 mM | 32.92 mL | 164.58 mL | 329.15 mL |
| 0.1 mM | 16.46 mL | 82.29 mL | 164.58 mL |
| 0.5 mM | 3.29 mL | 16.46 mL | 32.92 mL |
Calculators
Background References
References are publications that support the biological activity of the product. See our Citations tab to view 629 publications citing the usage of this product.
- Weichhart Inhibition of mTOR blocks the anti-inflammatory effects of glucocorticoids in myeloid immune cells. Blood 2011 PMID: 21368289
- Li mTORC1 inhibition increases neurotensin secretion and gene expression through activation of the MEK/ERK/c-Jun pathway in the human endocrine cell line BON. Am.J.Physiol.Cell Physiol. 2011 PMID: 21508335
- Galluzzi Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles. Nat.Rev.Drug.Discov. 2017 PMID: 28529316
- Peterson mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell. 2011 PMID: 21816276
- Liu Discovery of 1-(4(-(4-propionylpiperazin-1-yl)-3-(trifluoromethyl)phenyl)-9-(quinolin-3-yl)benzo[h][1,6]naphthyridin-2(1H)-one as a highly potent, selective mammalian target of rapamycin (mTOR) inhibitor for the treatment of cancer. J.Med.Chem. 2010 PMID: 20860370
- Thoreen An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J.Biol.Chem. 2009 PMID: 19150980
- Guertin and Sabatini The pharmacology of mTOR inhibition. Sci.Signal. 2009 PMID: 19383975
Product Documents for Torin 1
Certificate of Analysis
To download a Certificate of Analysis, please enter a lot or batch number in the search box below.
Product Specific Notices for Torin 1
For research use only
Related Research Areas
Citations for Torin 1
Customer Reviews for Torin 1 (2)
Have you used Torin 1?
Submit a review and receive an Amazon gift card!
$25/€18/£15/$25CAN/¥2500 Yen for a review with an image
$10/€7/£6/$10CAN/¥1110 Yen for a review without an image
Submit a review
Customer Images
-
Species: MouseAssay Type: In VitroCell Line/Tissue: Primary pancreatic ductal epithelial cellsVerified Customer | Posted 04/03/20200.076–500 nM Torin
-
Species: MouseAssay Type: Ex VivoCell Line/Tissue: acute brain slicesVerified Customer | Posted 01/10/201830 minute pre-incubation on acute slicesWe used Torin-1 to inhibit torin signalling, we got the expected effect on metabolic changes in acute brain slices
There are no reviews that match your criteria.

